Drosophila Developmental Neuroscience and Gene Regulation
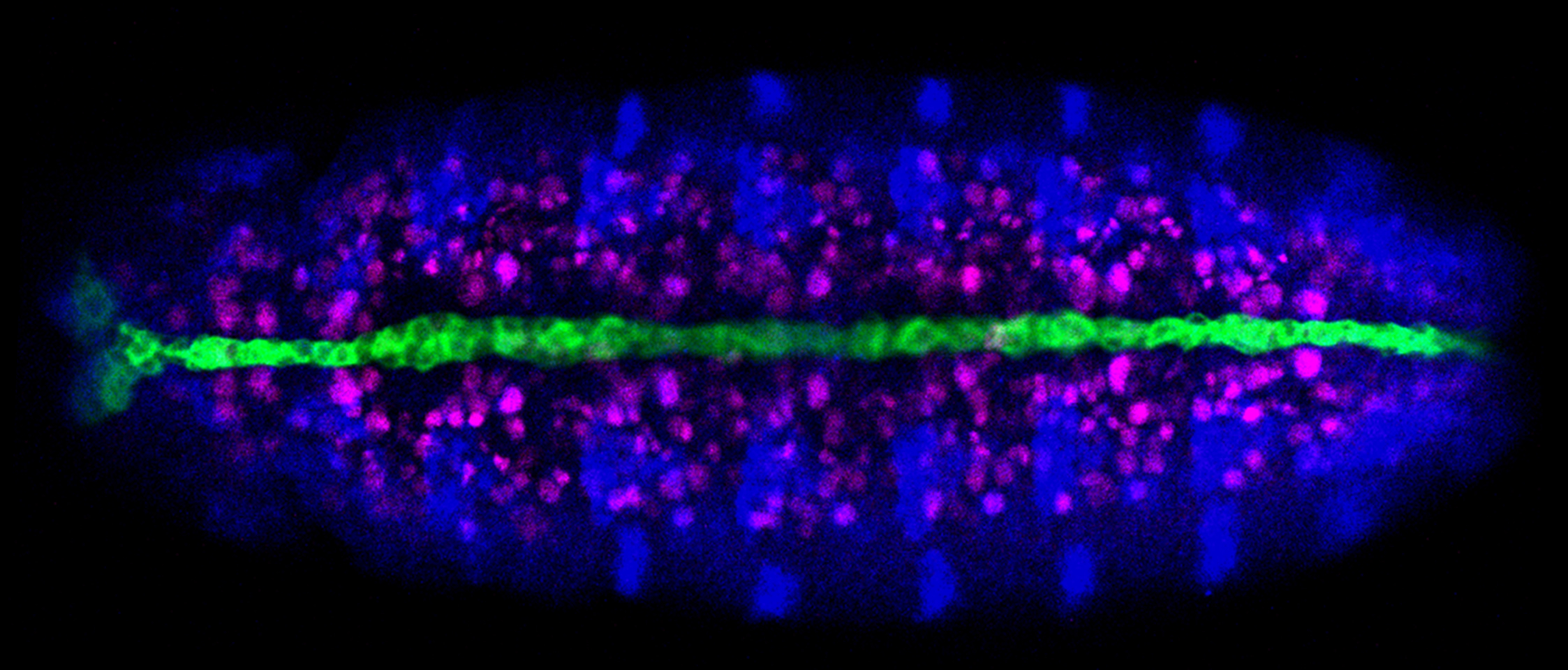
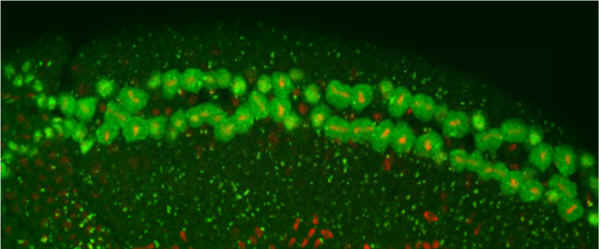
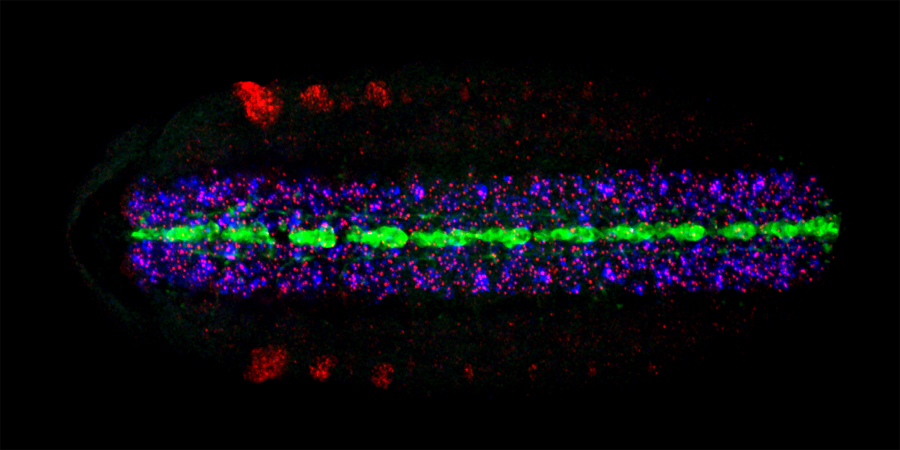
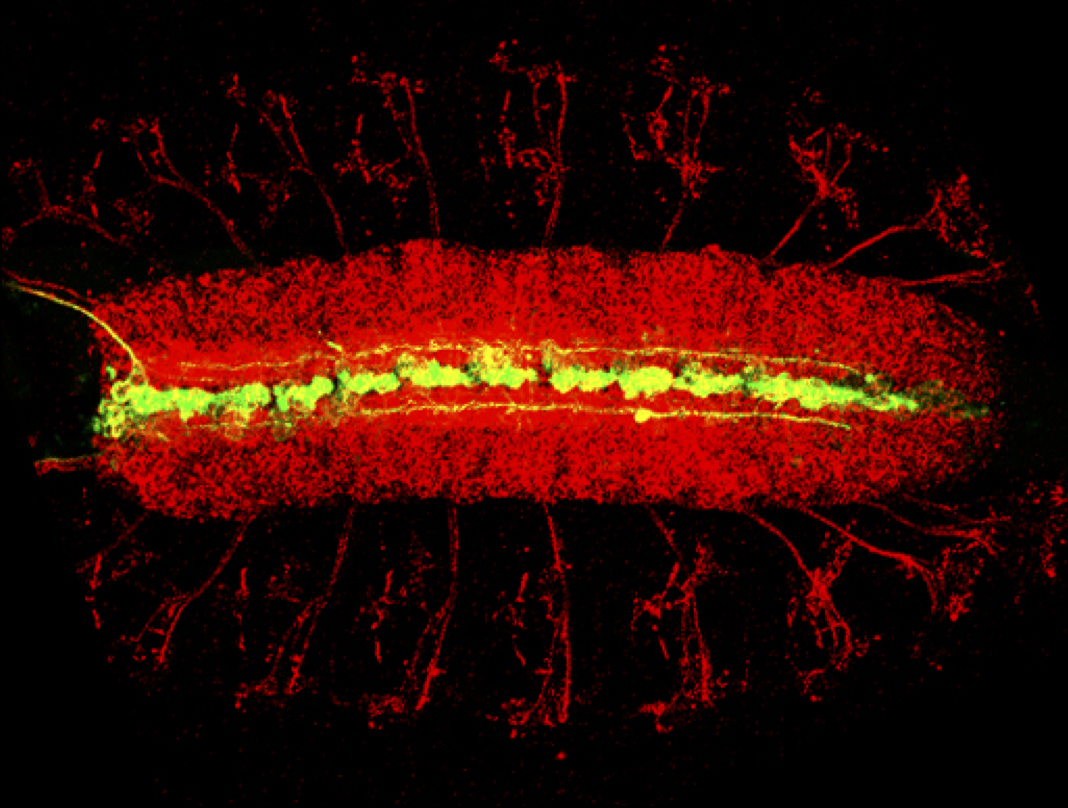
The long-term goal of our lab’s research program has been to understand the molecular basis of Drosophila CNS development. The experimental system we studied were the neurons and glia that reside along the midline of the embryonic CNS. Much of our early work studied the Drosophila single-minded bHLH-PAS transcription factor gene, the master regulator of CNS midline cell development. In recent years, we utilized genetic, molecular, genomic, and cellular approaches, including live imaging, to pioneer understanding of Drosophila CNS midline cell development. Our studies revealed the regulatory circuitry controlling neuronal and glial cell fate and differentiation, the properties of midline transcriptional enhancer elements, the control of neuronal and glial cell migration, and how axons and glia interact.
NO LONGER ACCEPTING RESEARCH STUDENTS